The MVB sorting pathway plays a crucial role in growth-factor-receptor downregulation1, developmental signaling2-4 and regulation of the immune response5. In eukaryotic cells, MVBs are formed after invagination of the limiting membrane of the sorting endosome, delivering transmembrane proteins and lipids to the lumen. Enveloped viruses such as human immunodeficiency virus hijack the MVB pathway in order to escape from cells5, 6.
Cargo sorting into MVB is a highly regulated process and it has been shown that monoubiquitination of transmembrane proteins serve as a signal for sorting into the MVB pathway7, 8. The vesicle formation and cargo sorting depends on a group of at least 20 proteins first identified in Baker's yeast, Saccharomyces cerevisiae, and known as class E Vacuolar Protein Sorting(vps) proteins9, 10. (The vacuole is the yeast counterpart of the lysosome). These vps proteins are highly conserved from yeast to humans with only the addition of a few more homologs in humans indicating a more regulated MVB sorting pathway. The majority of the class E vps proteins can be grouped in three separate heteromeric subcomplexes termed ESCRT-I, ESCRT-II and ESCRT-III (Endosomal Sorting Complex Required for Transport). During MVB sorting, HRS-vps27 is first recruited to the early endosome by virtue of its FYVE domain interaction with PI(3)P and its UIM (ubiquitin interacting motif) interaction with ubiquitinated cargo. It then recruits the ESCRT-I complex (composed of vps23, vps28 and vps37) to the membrane. ESCRT-I recruits the downstream ESCRT-II and ESCRT-III complexes11. After the ESCRTs have been recruited to the endosome membrane, the AAA-type ATPase vps4 binds ESCRT-III and following MVB vesicle formation catalyses the dissociation of ESCRT protein complexes in an ATP-dependent manner for further rounds of protein sorting12. Together, the ESCRT complexes, vps4, and related proteins form a complex membrane-associated network. Our laboratories have the long term goal of carrying out a complete three dimensional structural and functional characterization of this network.
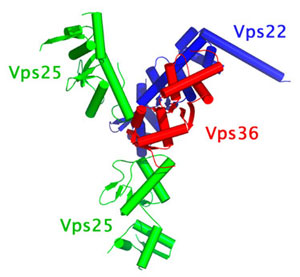
Using x-ray diffraction data collected at the APS and SSRL Beam Line 9-2 we have determined the first structure of an ESCRT complex. Here we report the 3.6 Å resolution structure of the quaternary ESCRT-II complex containing vps22, vps36 and two vps25 molecules. The complex adopts the shape of a capital letter 'Y' (figure 1), with overall dimensions of 120 × 85 × 52 Å. The base of the 'Y' consists of one of the two molecules of vps25; one of the branches of the 'Y' consists of the second molecule of vps25, and the other branch is formed by a subcomplex consisting of vps22 and the C-terminal domain of vps36. The N terminus of vps22 is a single a-helix that protrudes away from the tip of the 'Y' shape. None of the three ESCRT-II subunits has discernable homology between each other at the amino acid sequence level. However all three have in common a structure based on two copies of the winged-helix (WH) fold.
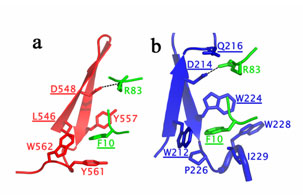
The vps22-vps36 interaction is the most extensive in the complex burying a total of 2,527 Å2 of solvent-accessible surface area from both partners. The vps22-vps25 interaction buries a total of 1,689 Å2 of solvent-accessible surface. The focal point of the vps25 binding site on vps22 is an aromatic cage made up of the side chains of Trp 212, Trp 224 and Trp 228. Pro 226 and Ile 229 also contribute to the walls of the cage. Together these residues completely surround and bury Phe 10 of vps25. In addition to hydrophobic interactions, a salt bridge is formed by the side chains of Asp 214 (vps22) and Arg 83 (vps25). The parallels between vps36-vps25 and vps22-vps25 interactions are striking (figure 2). Phe 10 of vps25 is buried in an aromatic cage on vps36. The cage is formed by Leu 546, Tyr 557, Tyr 561 and Trp 562. Also a salt bridge is formed between Arg 83 (vps25) and Asp 584 (vps36). The total surface area buried is 1,524 Å2. The mutation F10D at the heart of the vps25 interface with vps22 or vps36, or mutation R83D, with removes salt bridges with vps22 Asp 214 and vps36 Asp 548, leads in both cases to a class E phenotype and a monomer vps25. The salt bridge can be rescued by a charge reversal mutation recovering the stoichiometry and functionality of the complex.
The structure shows how ESCRT-II subunits assemble to form a scaffold to coordinate multiple interactions with proteins. As the first structure of ESCRT complex it is an exciting step forward. By the same token, much work remains to achieve the ultimate goal of a three dimensional mechanistic understanding at of the entire MVB protein network.
- Futter, C. E., Pearse, A., Hewlett, L. J. & Hopkins, C. R. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 132, 1011-1023 (1996).
- Deblandre, G. A., Lai, E. C. & Kintner, C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell 1, 795-806 (2001).
- Lai, E. C., Deblandre, G. A., Kintner, C. & Rubin, G. M. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of Delta. Dev. Cell 1, 783-794 (2001).
- Pavlopoulos, E. et al. Neuralized encodes a peripheral membrane protein involved in Delta signaling and endocytosis. Dev. Cell 1, 807-816 (2001).
- Kleijmeer, M. et al. Reorganization of multivesicular bodies regulates MHC class II antigen presentation by dendritic cells. J. Cell Biol. 155, 53-63 (2001).
- von Schwedler, U. K. et al. The protein network of HIV budding. Cell. 2003 Sep 19; 114(6):701-13.
- Katzmann, D. J., Babst, M. & Emr, S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145-155 (2001).
- Reggiori, F. & Pelham, H. R. B. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20, 5176-5186 (2001).
- Raymond, C. K., Roberts, C. J., Moore, K. E., Howald, I. & Stevens, T. H. Biogenesis of the vacuole in Saccharomyces cerevisiae. Int. Rev. Cytol. 139, 59-120 (1992).
- Katzmann, D. J., Odorizzi, G. & Emr, S. D. Receptor downregulation and multivesicular-body sorting. Nature Rev. Mol. Cell Biol. 3, 893-905 (2002).
- Babst, M., Katzmann, D. J., Snyder, W. B., Wendland, B. & Emr, S. D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283-289 (2002).
- Babst, M., Wendland, B., Estepa, E. J. & Emr, S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982-2993 (1998).
Hierro, A., Sun, J., Rusnak, A. S., Kim, J., Prag, G., Emr, S. D. & Hurley, J. H. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004 Sep 9; 431(2004):221-5.




