Clostridal neurotoxins (CNTs) are the causative agents of the neuroparalytic diseases botulism and tetanus1,2 CNTs impair neuronal exocytosis (a process by which neurotransmitter is released into a synapse) through specific proteolysis of essential proteins called SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors)3. SNARE assembly into a low-energy ternary complex is believed to catalyse membrane fusion, precipitating neurotransmitter release; this process is attenuated in response to SNARE proteolysis4-7. Site-specific SNARE hydrolysis is catalysed by the CNT light chains, a unique group of zinc-dependent endopeptidases (enzymes that catalyse hydrolysis of peptide bonds within other proteins)3. The means by which a CNT properly identifies and cleaves its target SNARE has been a subject of much speculation; it is thought to use one or more regions of enzyme-substrate interaction remote from the active site (exosites)8-10. Using x-ray diffraction data collected in part at SSRL beamline 9-2, we have determined the first structure of a CNT endopeptidase in complex with its target SNARE at a resolution of 2.1 Å: botulinum neurotoxin serotype A (BoNT/A) protease bound to human SNAP-25. The structure, together with enzyme kinetic data, reveals an array of exosites that determine substrate specificity (figure 1).
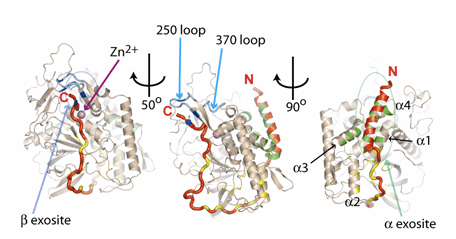
The α-exosite (indicated by a green arrow in Fig. 1) is formed by BoNT/A light-chain helices (tan) α1-α4 that bind to the helical N-terminus of the substrate (red). Green areas indicate the approximate locations of contacting side chains involved in the α-exosite. On the opposite face of BoNT/A, the β-exosite is indicated by a blue arrow (Fig. 1). The C-terminus of SNAP-25 forms an anti-parallel β sheet along with a portion of the '250 loop' (light-blue), which is separated from the active site (indicated by Zn2+, purple sphere) by the '370 loop' (light-blue). Dark-blue areas indicate the approximate locations of contacting side chains involved in the b-exosite. Yellow areas indicate the approximate locations of other exosites (anchor points) involved in side-chain contacts between the SNAP-25 substrate and the BoNT/A light chain.
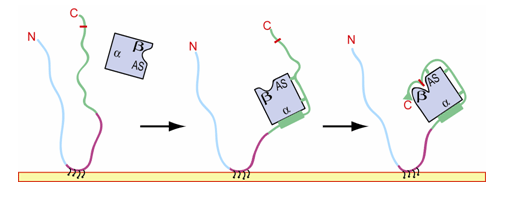
Based on our structure and available kinetic data for several mutant SNAP-25 substrates, we conclude that most of this unusually large enzyme-substrate interface serves to provide a substrate-specific boost to catalytic efficiency by reducing Km (the Michaelis constant). We also observe significant structural changes near the toxin's catalytic pocket upon substrate binding, probably serving to render the protease competent for catalysis. A general model of the strategy used by BoNT/A to recognize and cleave SNAP-25 is presented in figure 2. SNAP-25 is shown attached to a presynaptic membrane via palmitylation sites (shown in black) on its linker domain (purple). The N-terminal (sn1, cyan) and C-terminal (sn2, green) domains are unstructured or flexible in uncomplexed SNAP-25 (ref. 11). Binding of BoNT/A (blue) is probably initiated by helix formation at the α-exosite, and anchor points along the extended portion of SNAP-25 (green notches) are additional determinants of substrate specificity. These sites reduce Km and enhance binding at the β-exosite, inducing conformational changes at the active site, which render the endopeptidase competent to cleave its substrate.
Ultimately, the novel structures of the substrate-recognition exosites could be used for designing inhibitors specific to BoNT/A.
- Humeau, Y., Doussau, F., Grant, N. J. & Poulain, B. How botulinum and tetanus neurotoxins block neurotransmitter release. Biochimie 82, 427-446 (2000).
- Dolly, J. O., Black, J., Williams, R. S. & Melling, J. Acceptors for botulinum neurotoxin reside on motor nerve terminals and mediate its internalization. Nature 307, 457-460 (1984).
- Schiavo, G. et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 359, 832-835 (1992).
- Chen, Y. A., Scales, S. J., Patel, S. M., Doung, Y. C. & Scheller, R. H. SNARE complex formation is triggered by Ca2+ and drives membrane fusion. Cell 97, 165-174 (1999).
- Söllner, T. et al. SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318-324 (1993).
- Pellegrini, L. L., O'Connor, V., Lottspeich, F. & Betz, H. Clostridial neurotoxins compromise the stability of a low energy SNARE complex mediating NSF activation of synaptic vesicle fusion. EMBO J. 14, 4705-4713 (1995).
- Xu, T., Binz, T., Niemann, H. & Neher, E. Multiple kinetic components of exocytosis distinguished by neurotoxin sensitivity. Nature Neurosci. 1, 192-200 (1998).
- Rossetto, O. et al. SNARE motif and neurotoxins. Nature 372, 415-416 (1994).
- Pellizzari, R. et al. Structural determinants of the specificity for synaptic vesicle-associated membrane protein/synaptobrevin of tetanus and botulinum type B and G neurotoxins. J. Biol. Chem. 271, 20353-20358 (1996).
- Cornille, F. et al. Cooperative exosite-dependent cleavage of synaptobrevin by tetanus toxin light chain. J. Biol. Chem. 272, 3459-3464 (1997).
- Fasshauer, D., Bruns, D., Shen, B., Jahn, R. & Brunger, A. T. A structural change occurs upon binding of syntaxin to SNAP-25. J. Biol. Chem. 272, 4582-4590 (1997).
Breidenbach, M. A. & Brunger, A. T. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 432, 925-929 (2004)




