Nitrogen is a key element for building essential organic molecules required for life, such as proteins and nucleic acids. It is found abundantly as the dinitrogen (N2) gas in the atmosphere, making up 78% of the air we breathe. The strong triple-bond between the nitrogen atoms in N2 makes this compound difficult to be broken apart into usable forms within the body. However, certain bacteria are able to convert N2 into accessible forms of nitrogen that can be used to construct the building blocks for all living creatures, including humans. Only a select group of bacteria can perform this critical reaction because they produce the enzyme nitrogenase that is able to catalyze the conversion of the relatively inert N2 gas.
Nitrogenase is a complex enzyme that uses a suite of metallocofactors to catalyze the energetically demanding reduction of N2 to ammonia under ambient conditions. Many efforts have gone to understand the mechanisms of nitrogenase since its discovery, yet they have met with limited success. The research groups of Professors Hu and Ribbe at the University of California-Irvine applied the strategy to limit the electron supply for substrate turnover to capture a state of the nitrogenase enzyme bound with N2 or N2-derived intermediates for combined biochemical and structural analyses. Enhanced anomalous x-ray diffraction data were collected at the Fe absorption edge on BL12-2. The results of their analysis revealed that the three sulfur sites in the “belt region” of the active site cofactor (M-cluster) of nitrogenase are labile during catalysis, with the cofactor of one subunit of the enzyme having one, and the other subunit of the enzyme having two, of the three “belt sulfur” atoms replaced by distinct dinitrogen species during the binding and reduction of N2. These findings are entirely unexpected and shed important light on the sparsely understood mechanisms of N2 reduction, pointing to a key role of belt sulfur displacement in proper nitrogenase function.
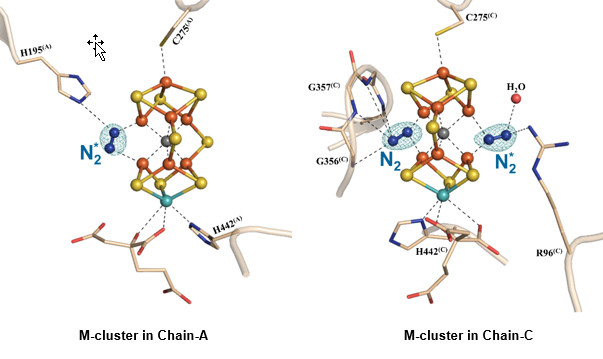
Figure 1. The nitrogen ligand-bound M-clusters in nitrogenase. Side view of M-clusters in Chain-A and Chain-C refined at a resolution of 1.83 Å. Key residues interacting with the clusters and the bound dinitrogen species (N2*) are shown in ball-and-stick presentations. The structural models of both clusters are superimposed with the Fo-Fc omit maps of the nitrogen ligands, contoured at 10 σ (mint-blue mesh). Color code of atoms: Fe, orange; Mo, aqua; S, yellow; O, red; N, blue.
The molecular depiction of the N2- or intermediate-bound state of nitrogenase has remained the central goal of nitrogenase research for nearly four decades. The structural snapshot reported by Professors Hu and Ribbe is crucial for understanding the strategy devised by nature for the binding and activation of the relatively inert dinitrogen gas to make the essential element nitrogen available for every living organism on earth. Moreover, given that ammonia is an excellent carbon-free fuel source, knowledge derived from this study could be instrumental in future development of nitrogenase-based biotechnological applications, which have profound implications for the energy- and environment-related areas.
W. Kang, C. C. Lee, A. J. Jasniewski, M. W. Ribbe and Y. Hu, "Structural Evidence for a Dynamic Metallocofactor during N2 Reduction by Mo-nitrogenase", Science 368, 1381 (2020) doi: 10.1126/science.aaz674

